Survival Analysis¶
Mining Excavator dataset case study
[1]:
from pathlib import Path
import numpy as np
import pandas as pd
import seaborn as sns
import matplotlib.pyplot as plt
%matplotlib inline
import nestor
from nestor import keyword as kex
import nestor.datasets as dat
def set_style():
# This sets reasonable defaults for font size for a figure that will go in a paper
sns.set_context("paper")
# Set the font to be serif, rather than sans
sns.set(font='serif')
# Make the background white, and specify the specific font family
sns.set_style("white", {
"font.family": "serif",
"font.serif": ["Times", "Palatino", "serif"]
})
set_style()
/home/tbsexton/anaconda3/envs/nestor-dev/lib/python3.6/importlib/_bootstrap.py:219: RuntimeWarning: numpy.dtype size changed, may indicate binary incompatibility. Expected 96, got 88
return f(*args, **kwds)
/home/tbsexton/anaconda3/envs/nestor-dev/lib/python3.6/importlib/_bootstrap.py:219: RuntimeWarning: numpy.dtype size changed, may indicate binary incompatibility. Expected 96, got 88
return f(*args, **kwds)
[2]:
df = dat.load_excavators()
df.head().style
[2]:
| BscStartDate | Asset | OriginalShorttext | PMType | Cost | |
|---|---|---|---|---|---|
| 0 | 2004-07-01 00:00:00 | A | BUCKET WON'T OPEN | PM01 | 183.05 |
| 1 | 2005-03-20 00:00:00 | A | L/H BUCKET CYL LEAKING. | PM01 | 407.4 |
| 2 | 2006-05-05 00:00:00 | A | SWAP BUCKET | PM01 | 0 |
| 3 | 2006-07-11 00:00:00 | A | FIT BUCKET TOOTH | PM01 | 0 |
| 4 | 2006-11-10 00:00:00 | A | REFIT BUCKET TOOTH | PM01 | 1157.27 |
Knowledge Extraction¶
Import vocabulary from tagging tool¶
[3]:
# merge and cleanse NLP-containing columns of the data
nlp_select = kex.NLPSelect(columns = ['OriginalShorttext'])
raw_text = nlp_select.transform(df)
[4]:
tex = kex.TokenExtractor()
toks = tex.fit_transform(raw_text)
#Import vocabulary
vocab_path = Path('.')/'support'/'mine_vocab_1g.csv'
vocab = kex.generate_vocabulary_df(tex, init=vocab_path)
tag_df = kex.tag_extractor(tex, raw_text, vocab_df=vocab)
relation_df = tag_df.loc[:, ['P I', 'S I']]
tags_read = kex._get_readable_tag_df(tag_df)
tag_df = tag_df.loc[:, ['I', 'P', 'S', 'U', 'X', 'NA']]
intialized successfully!
intialized successfully!
Quality of Extracted Keywords¶
[5]:
nbins = int(np.percentile(tag_df.sum(axis=1), 90))
print(f'Docs have at most {nbins} tokens (90th percentile)')
Docs have at most 5 tokens (90th percentile)
[6]:
tags_read.join(df[['OriginalShorttext']]).sample(10)
[6]:
| I | NA | P | S | U | X | OriginalShorttext | |
|---|---|---|---|---|---|---|---|
| 5033 | engine, light, bay | changeout | Eng bay lights u/s changeout | ||||
| 3813 | text, bolts | broken | broken bolts TEXT | ||||
| 2436 | line, steel, mcv | reseal | Reseal MCV Steel lines | ||||
| 1963 | hyd | error | repair | temp | REPAIR HYDRAULIC TEMP ERROR | ||
| 2020 | pump, hyd, valve | 1main | reseal | relief | reseal#1main hyd. pump relief valve. | ||
| 2105 | hose, control, mcv | replace | REPLACE MCV CONTROL HOSE. | ||||
| 4884 | right_hand, camera | working | RH CAMERA NOT WORKING | ||||
| 3896 | horn, bracket | fit, 2nd | mounting | make | Fit up 2nd horn & make mounting bracket | ||
| 5009 | light, rear, counterweight | replace | Replace rear counterweight lights x 2 | ||||
| 2996 | lube | fault | lube fault |
[7]:
# how many instances of each keyword class are there?
print('named entities: ')
print('I\tItem\nP\tProblem\nS\tSolution')
print('U\tUnknown\nX\tStop Word')
print('total tokens: ', vocab.NE.notna().sum())
print('total tags: ', vocab.groupby("NE").nunique().alias.sum())
vocab.groupby("NE").nunique()
named entities:
I Item
P Problem
S Solution
U Unknown
X Stop Word
total tokens: 1767
total tags: 492
[7]:
| NE | alias | notes | score | |
|---|---|---|---|---|
| NE | ||||
| 1 | 3 | 2 | 766 | |
| I | 1 | 317 | 19 | 585 |
| P | 1 | 53 | 6 | 119 |
| S | 1 | 42 | 2 | 95 |
| U | 1 | 68 | 57 | 92 |
| X | 1 | 9 | 1 | 9 |
Effectiveness of Tags¶
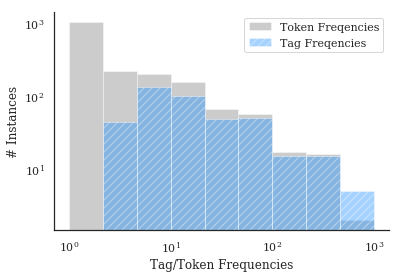
The entire goal, in some sense, is for us to remove low-occurence, unimportant information from our data, and form concept conglomerates that allow more useful statistical inferences to be made. Tags from nestor-gui, as the next plot shows, have no instances of 1x-occurrence concepts, compared to several thousand in the raw-tokens (this is by design, of course). Additionally, high occurence concepts that might have had misspellings or synonyms drastically inprove their average occurence
rate.
[8]:
cts = (tex._model.transform(raw_text)>0.).astype(int).toarray().sum(axis=0)
# cts2 = (tex3._model.transform(replaced_text2)>0.).astype(int).toarray().sum(axis=0)
sns.distplot(cts,
# np.concatenate((cts, cts2)),
bins=np.logspace(0,3,10),
# bins=np.linspace(0,1500,10),
norm_hist=False,
kde=False,
label='Token Freqencies',
hist_kws={'color':'grey'})
# cts
sns.distplot(tag_df[['I', 'P', 'S']].sum(),
bins=np.logspace(0,3,10),
# bins=np.linspace(0,1500,10),
norm_hist=False,
kde=False,
label='Tag Freqencies',
hist_kws={'hatch':'///', 'color':'dodgerblue'})
plt.yscale('log')
plt.xscale('log')
tag_df.sum().shape, cts.shape
plt.legend()
plt.xlabel('Tag/Token Frequencies')
plt.ylabel('# Instances')
sns.despine()
plt.savefig('toks_v_tags.png', dpi=300)
[9]:
# tag-completeness of work-orders?
tag_pct, tag_comp, tag_empt = kex.get_tag_completeness(tag_df)
# with sns.axes_style('ticks') as style:
sns.distplot(tag_pct.dropna(),
kde=False, bins=nbins,
kde_kws={'cut':0})
plt.xlim(0.1, 1.0)
plt.xlabel('precision (PPV)')
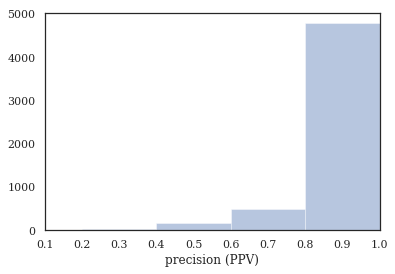
Tag completeness: 0.94 +/- 0.13
Complete Docs: 4444, or 81.02%
Empty Docs: 48, or 0.88%
[9]:
Text(0.5,0,'precision (PPV)')
Convergence over time, using nestor-gui¶
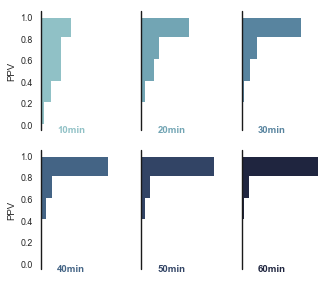
As part of the comparison study, an expert used nestor-gui for approximately 60min annotating 1-grams, followed by 20min focusing on 2-grams. Work was saved every 10 min, so we would like to see how the above plot was arrived at as the tokens were classified.
[10]:
study_fname = Path('.')/'support'/'vocab_study_results.csv'
study_df = pd.read_csv(study_fname, index_col=0)
study_long = pd.melt(study_df, var_name="time", value_name='PPV').dropna()
study_long['time_val'] = study_long.time.str.replace('min','').astype(float)
sns.set(style="white", rc={"axes.facecolor": (0, 0, 0, 0)}, context='paper')
pal = sns.cubehelix_palette(6, rot=-.25, light=.7)
g = sns.FacetGrid(study_long, col="time", hue="time", aspect=.8, height=2, palette=pal, col_wrap=3)
g.map(sns.distplot, "PPV", kde=False, bins=nbins, vertical=True,
hist_kws=dict(alpha=1., histtype='stepfilled', edgecolor='w', lw=2))
g.map(plt.axvline, x=0, lw=1.4, clip_on=False, color='k')
# Define and use a simple function to label the plot in axes coordinates
def label(x, color, label):
ax = plt.gca()
ax.text(.2, 0, label, fontweight="bold", color=color,
ha="left", va="center", transform=ax.transAxes)
g.map(label, "PPV")
# Remove axes details that don't play well with overlap
g.set_titles("")
g.set( xticks=[], xlabel='')
g.set_axis_labels(y_var='PPV')
g.despine(bottom=True, left=True)
plt.tight_layout()
Survival Analysis¶
Rules-Based¶
From Hodkeiwiz et al, a rule-based method was used to estimate failure times for SA. Let’s see their data:
[9]:
df_clean = dat.load_excavators(cleaned=True)
df_clean['SuspSugg'] = pd.to_numeric(df_clean['SuspSugg'], errors='coerce')
df_clean.dropna(subset=['RunningTime', 'SuspSugg'], inplace=True)
df_clean.shape
[9]:
(5288, 17)
[10]:
df_clean.sort_values('BscStartDate').head(10)
[10]:
| BscStartDate | Asset | OriginalShorttext | PMType | Cost | RunningTime | MajorSystem | Part | Action | Variant | FM | Location | Comments | FuncLocation | SuspSugg | Rule | Unnamed: 16 | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 8 | 2001-07-19 | B | REPLACE LIP | PM01 | 1251.52 | 7.0 | Bucket | NaN | Replace | 2V | NaN | NaN | NaN | Bucket | 0.0 | Rule_1_3_78_383_384 | NaN |
| 1820 | 2001-09-01 | B | OIL LEAK L/H TRACK TENSIONER. | PM01 | 0.00 | 3.0 | Hydraulic System | Track | Minor Maint | 18 | Leak | Left | NaN | Power Train - Transmission | 0.0 | Rule_1_3_52_289_347_425_500 | NaN |
| 1821 | 2001-09-04 | B | BAD SOS METAL IN OIL | PM01 | 0.00 | 3.0 | Hydraulic System | Slew Gearbox | NaN | NaN | Contamination | NaN | NaN | Sprocket/Drive Compartment Right | 0.0 | Rule_1_3_52_303_409 | NaN |
| 5253 | 2001-09-05 | B | REPLACE AIRCONDITIONER BELTS | PM01 | 0.00 | 23.0 | NaN | Air Conditioning | Replace | 2V | NaN | NaN | NaN | Air Conditioning System | 0.0 | Rule_1_3_224_227_383_384 | NaN |
| 3701 | 2001-09-05 | B | REPLACE CLAMPS ON CLAM PIPES | PM01 | 0.00 | 28.0 | NaN | Mount | Replace | 2V | NaN | NaN | NaN | Oil - Hydraulic | 0.0 | Rule_1_3_92_181_383_384 | NaN |
| 1167 | 2001-09-05 | B | REPLACE RHS FAN BELT TENSIONER PULLEY | PM01 | 82.09 | 0.0 | NaN | Fan | Minor Maint_Replace | 2V | NaN | Right | NaN | +Cooling System | 0.0 | Rule_1_3_125_347_383_384_509 | NaN |
| 1168 | 2001-09-11 | B | replace fan belt | PM01 | 0.00 | 6.0 | NaN | Fan | Replace | 2V | NaN | NaN | NaN | +Cooling System | 0.0 | Rule_1_3_125_383_384 | NaN |
| 644 | 2001-09-15 | B | replace heads on lhs eng | PM01 | 0.00 | 33.0 | Engine | NaN | Replace | 2V | NaN | Left | NaN | Engine Left Cylinder Heads | 0.0 | Rule_1_3_25_383_384_499 | NaN |
| 4583 | 2001-09-26 | B | REPAIR CABIN DOOR FALLING OFF. | PM01 | 0.00 | 27.0 | NaN | Drivers Cabin | Repair | 1 | NaN | NaN | NaN | Operators Cabin | 0.0 | Rule_1_3_251_284_357 | NaN |
| 9 | 2001-10-01 | B | rebuild lip #3 | PM01 | 0.00 | 74.0 | Bucket | NaN | Repair | 5 | NaN | NaN | NaN | Bucket Clam (Lip) | 0.0 | Rule_1_3_78_362 | NaN |
We once again turn to the library Lifelines as the work-horse for finding the Survival function.
[11]:
from lifelines import WeibullFitter, ExponentialFitter, KaplanMeierFitter
mask = (df_clean.MajorSystem =='Bucket')
# mask=df_clean.index
def mask_to_ETclean(df_clean, mask, fill_null=1.):
filter_df = df_clean.loc[mask]
g = filter_df.sort_values('BscStartDate').groupby('Asset')
T = g['BscStartDate'].transform(pd.Series.diff).dt.days
# T.loc[(T<=0.)|(T.isna())] = fill_null
E = (~filter_df['SuspSugg'].astype(bool)).astype(int)
return T.loc[~((T<=0.)|(T.isna()))], E.loc[~((T<=0.)|(T.isna()))]
T, E = mask_to_ETclean(df_clean, mask)
wf = WeibullFitter()
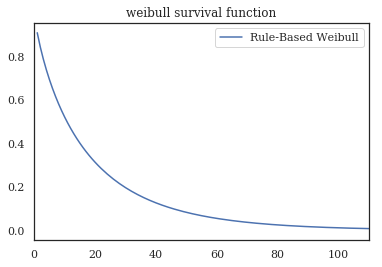
wf.fit(T, E, label='Rule-Based Weibull')
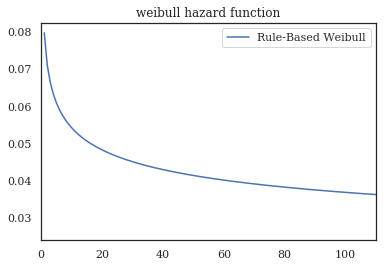
print('{:.3f}'.format(wf.lambda_), '{:.3f}'.format(wf.rho_))
# wf.print_summary()
wf.hazard_.plot()
plt.title('weibull hazard function')
plt.xlim(0,110)
wf.survival_function_.plot()
plt.xlim(0,110)
plt.title('weibull survival function')
print(f'transform: β={wf.rho_:.2f}\tη={1/wf.lambda_:.2f}')
# wf._compute_standard_errors()
to_bounds = lambda row:'±'.join([f'{i:.2g}' for i in row])
wf.summary.iloc[:,:2].apply(to_bounds, 1)
0.060 0.833
transform: β=0.83 η=16.73
[11]:
lambda_ 0.06±0.0033
rho_ 0.83±0.026
dtype: object
Tag Based Comparison¶
We estimate the occurence of failures with tag occurrences.
[ ]:
import math
def to_precision(x,p):
"""
returns a string representation of x formatted with a precision of p
Based on the webkit javascript implementation taken from here:
https://code.google.com/p/webkit-mirror/source/browse/JavaScriptCore/kjs/number_object.cpp
"""
x = float(x)
if x == 0.:
return "0." + "0"*(p-1)
out = []
if x < 0:
out.append("-")
x = -x
e = int(math.log10(x))
tens = math.pow(10, e - p + 1)
n = math.floor(x/tens)
if n < math.pow(10, p - 1):
e = e -1
tens = math.pow(10, e - p+1)
n = math.floor(x / tens)
if abs((n + 1.) * tens - x) <= abs(n * tens -x):
n = n + 1
if n >= math.pow(10,p):
n = n / 10.
e = e + 1
m = "%.*g" % (p, n)
if e < -2 or e >= p:
out.append(m[0])
if p > 1:
out.append(".")
out.extend(m[1:p])
out.append('e')
if e > 0:
out.append("+")
out.append(str(e))
elif e == (p -1):
out.append(m)
elif e >= 0:
out.append(m[:e+1])
if e+1 < len(m):
out.append(".")
out.extend(m[e+1:])
else:
out.append("0.")
out.extend(["0"]*-(e+1))
out.append(m)
return "".join(out)
[12]:
def query_experiment(name, df, df_clean, rule, tag, multi_tag, prnt=False):
def mask_to_ETclean(df_clean, mask, fill_null=1.):
filter_df = df_clean.loc[mask]
g = filter_df.sort_values('BscStartDate').groupby('Asset')
T = g['BscStartDate'].transform(pd.Series.diff).dt.days
E = (~filter_df['SuspSugg'].astype(bool)).astype(int)
return T.loc[~((T<=0.)|(T.isna()))], E.loc[~((T<=0.)|(T.isna()))]
def mask_to_ETraw(df_clean, mask, fill_null=1.):
filter_df = df_clean.loc[mask]
g = filter_df.sort_values('BscStartDate').groupby('Asset')
T = g['BscStartDate'].transform(pd.Series.diff).dt.days
T_defined = (T>0.)|T.notna()
T = T[T_defined]
# assume censored when parts replaced (changeout)
E = (~(tag_df.S.changeout>0)).astype(int)[mask]
E = E[T_defined]
return T.loc[~((T<=0.)|(T.isna()))], E.loc[~((T<=0.)|(T.isna()))]
experiment = {
'rules-based': {
'query': rule,
'func': mask_to_ETclean,
'mask': (df_clean.MajorSystem == rule),
'data': df_clean
},
'single-tag': {
'query': tag,
'func': mask_to_ETraw,
'mask': tag_df.I[tag].sum(axis=1)>0,
'data': df
},
'multi-tag': {
'query': multi_tag,
'func': mask_to_ETraw,
'mask': tag_df.I[multi_tag].sum(axis=1)>0,
'data': df
}
}
results = {
('query', 'text/tag'): [],
# ('Weibull Params', r'$\lambda$'): [],
('Weibull Params', r'$\beta$'): [],
('Weibull Params', '$\eta$'): [],
('MTTF', 'Weib.'): [],
('MTTF', 'K-M'): []
}
idx = []
for key, info in experiment.items():
idx += [key]
results[('query','text/tag')] += [info['query']]
if prnt:
print('{}: {}'.format(key, info['query']))
info['T'], info['E'] = info['func'](info['data'], info['mask'])
wf = WeibullFitter()
wf.fit(info['T'], info['E'], label=f'{key} weibull')
to_bounds = lambda row:'$\pm$'.join([to_precision(row[0],2),
to_precision(row[1],1)])
params = wf.summary.T.iloc[:2]
params['eta_'] = [1/params.lambda_['coef'], # err. propagation
(params.lambda_['se(coef)']/params.lambda_['coef']**2)]
params = params.T.apply(to_bounds, 1)
results[('Weibull Params', r'$\eta$')] += [params['eta_']]
results[('Weibull Params', r'$\beta$')] += [params['rho_']]
if prnt:
print('\tWeibull Params:\n',
'\t\tη = {}\t'.format(params['eta_']),
'β = {}'.format(params['rho_']))
kmf = KaplanMeierFitter()
kmf.fit(info["T"], event_observed=info['E'], label=f'{key} kaplan-meier')
results[('MTTF','Weib.')] += [to_precision(wf.median_,3)]
results[('MTTF','K-M')] += [to_precision(kmf.median_,3)]
if prnt:
print(f'\tMTTF: \n\t\tWeib \t'+to_precision(wf.median_,3)+\
'\n\t\tKM \t'+to_precision(kmf.median_,3))
info['kmf'] = kmf
info['wf'] = wf
return experiment, pd.DataFrame(results, index=pd.Index(idx, name=name))
[14]:
bucket_exp, bucket_res = query_experiment('Bucket', df, df_clean,
'Bucket',
['bucket'],
['bucket', 'tooth', 'lip', 'pin']);
[15]:
tags = ['hyd', 'hose', 'pump', 'compressor']
hyd_exp, hyd_res = query_experiment('Hydraulic System', df, df_clean,
'Hydraulic System',
['hyd'],
tags)
[16]:
eng_exp, eng_res = query_experiment('Engine', df, df_clean,
'Engine',
['engine'],
['engine', 'filter', 'fan'])
[17]:
frames = [bucket_res, hyd_res, eng_res]
res = pd.concat(frames, keys = [i.index.name for i in frames],
names=['Major System', 'method'])
res
[17]:
| query | Weibull Params | MTTF | ||||
|---|---|---|---|---|---|---|
| text/tag | $\beta$ | $\eta$ | Weib. | K-M | ||
| Major System | method | |||||
| Bucket | rules-based | Bucket | 0.83$\pm$0.03 | 17$\pm$0.9 | 10.8 | 9.00 |
| single-tag | [bucket] | 0.83$\pm$0.03 | 27$\pm$2 | 17.1 | 15.0 | |
| multi-tag | [bucket, tooth, lip, pin] | 0.82$\pm$0.02 | 16$\pm$0.9 | 10.5 | 9.00 | |
| Hydraulic System | rules-based | Hydraulic System | 0.86$\pm$0.02 | 14$\pm$0.6 | 9.07 | 8.00 |
| single-tag | [hyd] | 0.89$\pm$0.04 | 36$\pm$3 | 24.1 | 25.0 | |
| multi-tag | [hyd, hose, pump, compressor] | 0.89$\pm$0.02 | 15$\pm$0.7 | 9.74 | 9.00 | |
| Engine | rules-based | Engine | 0.81$\pm$0.02 | 17$\pm$1 | 10.8 | 9.00 |
| single-tag | [engine] | 0.79$\pm$0.03 | 19$\pm$1 | 11.8 | 10.0 | |
| multi-tag | [engine, filter, fan] | 0.81$\pm$0.02 | 15$\pm$0.8 | 9.31 | 8.00 | |
[18]:
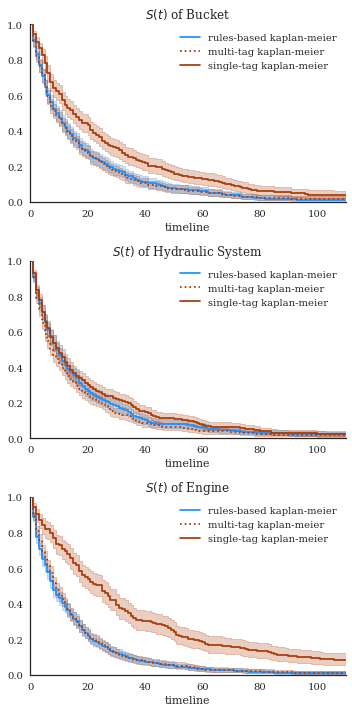
exp = [bucket_exp, eng_exp, hyd_exp]
f,axes = plt.subplots(nrows=3, figsize=(5,10))
for n, ax in enumerate(axes):
exp[n]['rules-based']['kmf'].plot(ax=ax, color='dodgerblue')
exp[n]['multi-tag']['kmf'].plot(ax=ax, color='xkcd:rust', ls=':')
exp[n]['single-tag']['kmf'].plot(ax=ax, color='xkcd:rust')
ax.set_xlim(0,110)
ax.set_ylim(0,1)
ax.set_title(r"$S(t)$"+f" of {res.index.levels[0][n]}")
sns.despine()
plt.tight_layout()
f.savefig('bkt_KMsurvival.png')
/home/tbsexton/anaconda3/lib/python3.6/site-packages/matplotlib/cbook/__init__.py:2446: UserWarning: Saw kwargs ['c', 'color'] which are all aliases for 'color'. Kept value from 'color'
seen=seen, canon=canonical, used=seen[-1]))
/home/tbsexton/anaconda3/lib/python3.6/site-packages/matplotlib/cbook/__init__.py:2446: UserWarning: Saw kwargs ['c', 'color'] which are all aliases for 'color'. Kept value from 'color'
seen=seen, canon=canonical, used=seen[-1]))
/home/tbsexton/anaconda3/lib/python3.6/site-packages/matplotlib/cbook/__init__.py:2446: UserWarning: Saw kwargs ['c', 'color'] which are all aliases for 'color'. Kept value from 'color'
seen=seen, canon=canonical, used=seen[-1]))
/home/tbsexton/anaconda3/lib/python3.6/site-packages/matplotlib/cbook/__init__.py:2446: UserWarning: Saw kwargs ['c', 'color'] which are all aliases for 'color'. Kept value from 'color'
seen=seen, canon=canonical, used=seen[-1]))
/home/tbsexton/anaconda3/lib/python3.6/site-packages/matplotlib/cbook/__init__.py:2446: UserWarning: Saw kwargs ['c', 'color'] which are all aliases for 'color'. Kept value from 'color'
seen=seen, canon=canonical, used=seen[-1]))
/home/tbsexton/anaconda3/lib/python3.6/site-packages/matplotlib/cbook/__init__.py:2446: UserWarning: Saw kwargs ['c', 'color'] which are all aliases for 'color'. Kept value from 'color'
seen=seen, canon=canonical, used=seen[-1]))
/home/tbsexton/anaconda3/lib/python3.6/site-packages/matplotlib/cbook/__init__.py:2446: UserWarning: Saw kwargs ['c', 'color'] which are all aliases for 'color'. Kept value from 'color'
seen=seen, canon=canonical, used=seen[-1]))
/home/tbsexton/anaconda3/lib/python3.6/site-packages/matplotlib/cbook/__init__.py:2446: UserWarning: Saw kwargs ['c', 'color'] which are all aliases for 'color'. Kept value from 'color'
seen=seen, canon=canonical, used=seen[-1]))
/home/tbsexton/anaconda3/lib/python3.6/site-packages/matplotlib/cbook/__init__.py:2446: UserWarning: Saw kwargs ['c', 'color'] which are all aliases for 'color'. Kept value from 'color'
seen=seen, canon=canonical, used=seen[-1]))
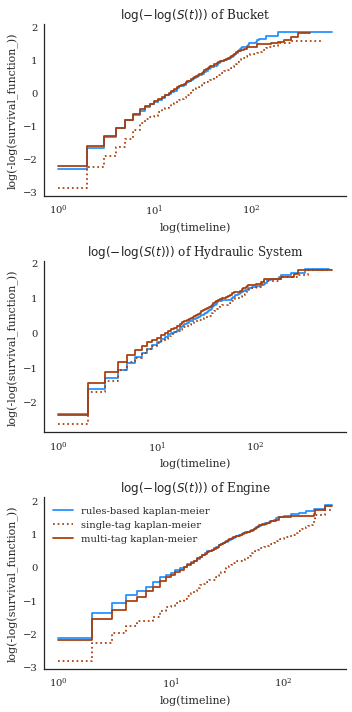
This next one give you an idea of the differences better. using a log-transform. the tags under-estimate death rates a little in the 80-130 day range, probably because there’s a failure mode not captured by the [bucket, lip, tooth] tags (because it’s rare).
[20]:
f,axes = plt.subplots(nrows=3, figsize=(5,10))
for n, ax in enumerate(axes):
exp[n]['rules-based']['kmf'].plot_loglogs(ax=ax, c='dodgerblue')
exp[n]['single-tag']['kmf'].plot_loglogs(ax=ax, c='xkcd:rust', ls=':')
exp[n]['multi-tag']['kmf'].plot_loglogs(ax=ax, c='xkcd:rust')
if n != 2:
ax.legend_.remove()
# ax.set_xlim(0,110)
# ax.set_ylim(0,1)
ax.set_title(r"$\log(-\log(S(t)))$"+f" of {res.index.levels[0][n]}")
sns.despine()
plt.tight_layout()
f.savefig('bkt_logKMsurvival.png', dpi=300)
# kmf.plot_loglogs()
[ ]: